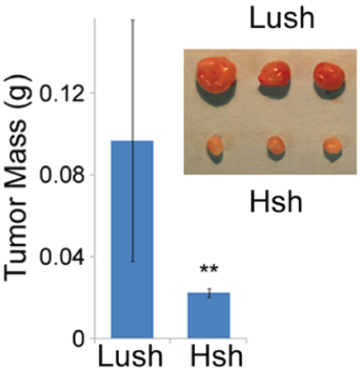
E3 Ubiquitin ligases have been very attractive therapeutic targets in cancer treatment, due to their major biological roles, and target specificity. HECTD3 is a novel HECT-type E3 ubiquitin (Ub) ligase expressed predominantly in T cells of the immune system and breast cancer cells. In collaboration with Drs. Ceshi Chen and Avram Dorina, we observed that HECTD3 stabilized MALT1 through polyubiquitination. Depletion of HECTD3, and the resulting reduction in levels of MALT1, had two major effects. First, cancer cells deficient in HECTD3 became more vulnerable to apoptosis induced by cisplatin, a chemotherapeutic agent. Second, HECTD3 deficient mice displayed reduced levels of MALT1 in isolated Th17 cells, reduced effector cytokine expression, and greatly diminished EAE severity, correlating with observations made in MALT1 knockout mice. These results strongly indicate that HECTD3 is a critical modulator of MALT1-dependent signaling. However, the exact mechanism of action of HECTD3 on MALT1 is still poorly understood. The goal of this project is to determine the structure and function of the HECTD3 ligase.
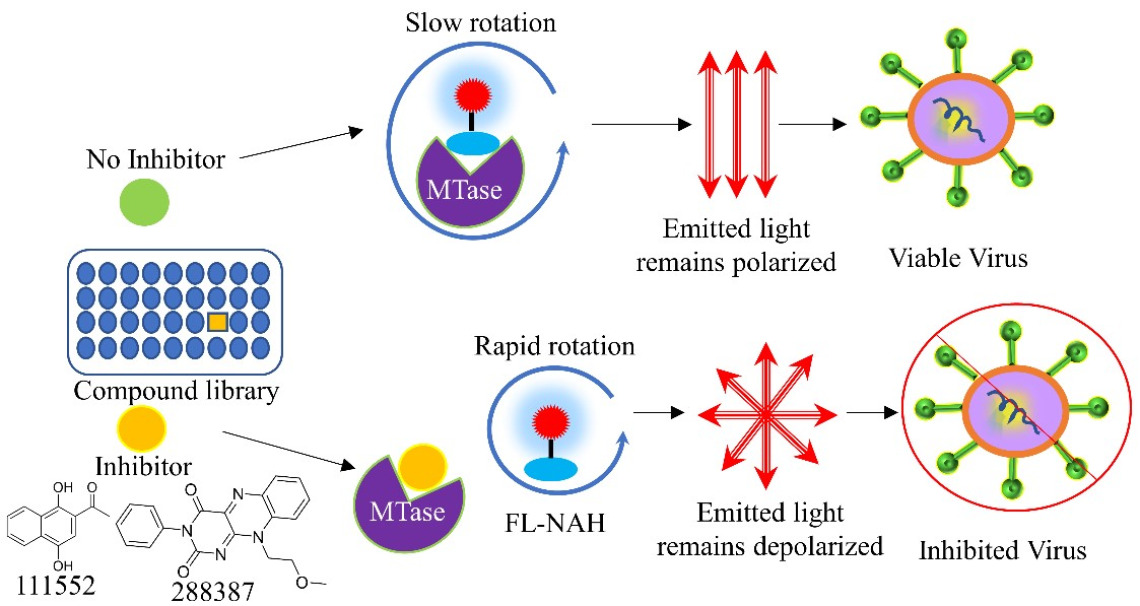
Large outbreaks or global pandemics of emerging and re-emerging viruses have occurred frequently in the past two decades with the most devasting being the current SARS-CoV-2 pandemic. Although three drugs were approved by FDA to treat SARS-CoV-2 infection, therapeutic agents against viruses are limited in general. Except for HIV and HCV that cause chronic infection, development of antivirals for acute viral infections was historically ignored until COVID-19. However, we should learn from previously missed opportunities. Coronaviruses (CoVs) such as NL63, OC43 and 229E are well known to only cause common cold, until outbreaks of genetically similar but lethal SARS-CoV2003, MERS-CoV2012, and SARS-CoV-22019. When SARS2003 disappeared, research in drug development against SARS and CoVs was generally halted due to lack of interest and difficulty in obtaining research funding. If the research had continued, we could have been better prepared for the current COVID-19 pandemic. Many viruses, particularly RNA viruses, are similar to CoVs that mutate or evolve frequently. They have pandemic potential. It is currently unclear what is going to be next outbreak.
Many viruses, including CoVs, ssRNA flaviviruses (Dengue virus serotypes 1-4 [DENV1-4], Zika virus [ZIKV], etc.), dsRNA reoviruses, dsDNA poxviruses, and (-)RNA viruses such as Henipaviruses encode their own MTases. It has been shown that viral N-7 MTase activity is essential for viral replication, making it an attractive drug target. We recently developed a universal HTS assay which is suitable for screening inhibitors of any SAM-dependent viral MTases. The assay is based on FL-NAH, a fluorescent analog of SAM and its reaction byproduct SAH. Our current goal is to screen, identify, and characterize novel broad-spectrum small molecule inhibitors against the viral methyltransferase, using both in vitro assays and in vivo animal models against representative viruses including but not limited to flaviviruses, alphaviruses, CoVs, Henipaviruses, poxviruses, and Ebola virus.
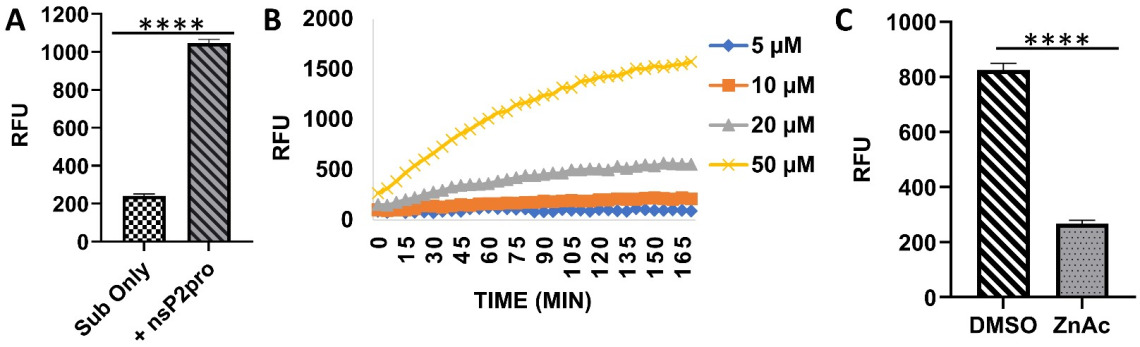
Alphavirus is the sole genus of RNA viruses in the Togaviridae family. Alphaviruses are arthropod-borne viruses with a positive-sense, single-stranded RNA genome. There are 32 alphaviruses including Chikungunya virus (CHIKV) and Western equine encephalitis virus (W EEV), Venezuelan EEV (VEEV) and Eastern EEV (EEEV) which is a select agent. Alphaviruses are generally spread by mosquitos, such as Aedes aegypti and A. albopictus. Infection of alphaviruses can cause human diseases ranging from mild rashes, fever, acute and chronic arthritis, to lethal encephalitis. Alphaviruses have caused several outbreaks, including CHIKV during 2013-14, when more than a million people across America contracted the virus. Currently, no vaccine or antiviral drug is available against CHIKV. In this project, we aim to target the alphavirus nonstructural protein 2 (nsP2), which has cysteine protease activity at its C-terminal domain. The nsP2 protease is essential for cleavage of the viral polyprotein precursor nsP1234 into functional nsPs. Thus, the nsP2 protease is an ideal target to develop direct acting antivirals (DAAs) to combat alphavirus infections. We will combine high-throughput screening (HTS) assays, sensitive biochemical assays, mass spectrometry (MS)-based functional assays, and cell-based antiviral assays to rapidly identify potent nsP2pro inhibitors to combat alphavirus infections.
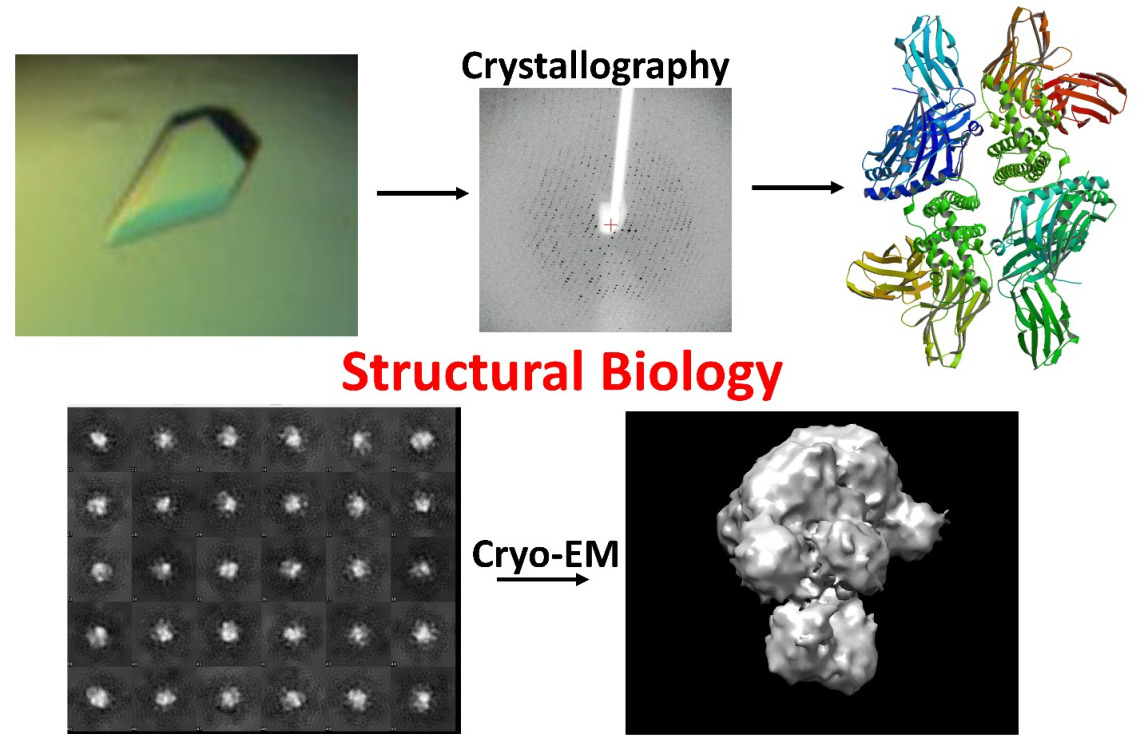
Understanding protein structure/function relationships is fundamental for our understanding of the biological process, and to provide new avenues for therapeutic invention. Although the recent development of AlphaFold2 has increased the accuracy of predicting protein structures, AlphaFold2 has limitations. Protein structures are more than just 3D atomic positions and can interact with each other to form complexes. They frequently adopt multiple conformations, have disordered regions, and may have flexible loops. Frequently, the enzyme active sites display significantly different conformations and may even disorder when not bound to ligand, substrate, or co-factor. AlphaFold2 was not designed for these purposes nor for prediction of how small molecules such as ligands, substrates, co-factors, and inhibitors bind to the enzyme active site. In contrast, precise structures are needed for structure-based drug discovery. Therefore, crystal structures at high resolution are still needed in the AlphaFold2 era.
Our laboratory's primary focus is to unravel the structures of crucial enzymes and proteins that play pivotal roles in various human diseases. Our research encompasses a wide range of conditions, such as Alzheimer's disease, breast cancer, multiple sclerosis, and infectious diseases caused by viruses, fungi, and mycobacteria. To achieve these goals, we employ cutting-edge structural biology techniques, including X-ray crystallography and Cryo-EM. Through these advanced approaches, we aim to gain critical insights into the molecular architecture of these disease-associated proteins and drug targets, paving the way for the development of targeted therapeutic interventions.
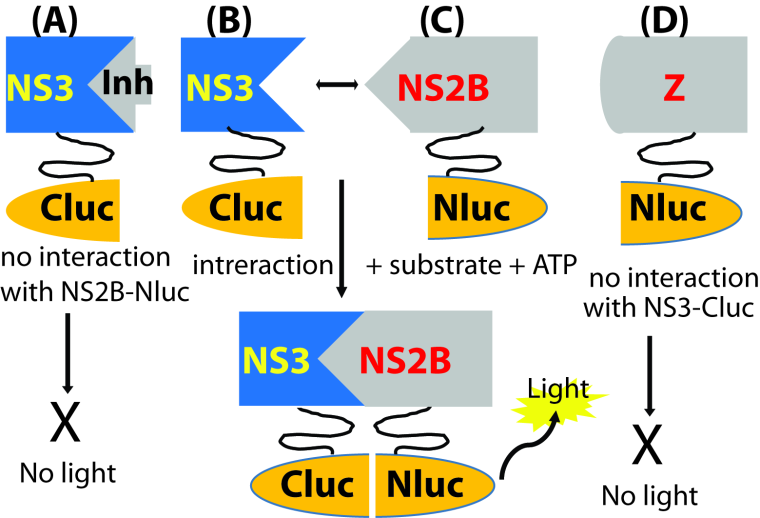
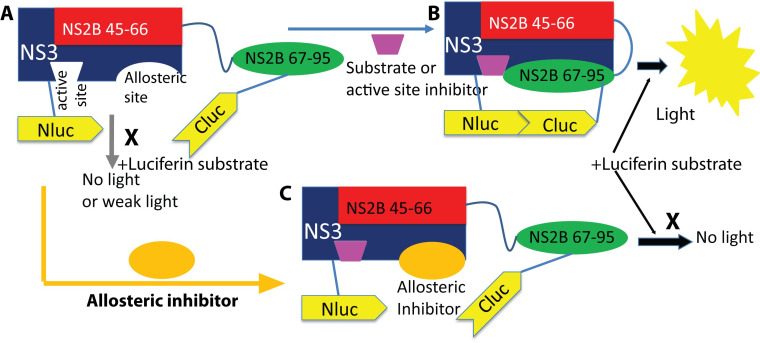
Many flaviviruses cause significant human diseases. However, no clinically approved antiviral therapy is available for treatment of flavivirus infections. Therefore, the development of vaccines and antiviral agents for prevention and treatment of flavivirus infections is a clear public health priority. The flavivirus NS2B-NS3 protease is an enzyme essential for viral polyprotein processing and maturation. We have developed high throughput screening assays based on split-luciferase complementation technology. We used these assays to screening inhibitors either blocking interactions between viral NS3 protease and its cofactor or preventing conformational changes of the cofactor NS2B. We identified several existing drugs that are potent and broad-spectrum inhibitors against several flaviviruses including Zika virus, Dengue virus, Yellow fever virus, Japanese encephalitis virus, West Nile virus, St. Louis encephalitis virus, and Powassan virus. We have shown that compound interfering NS2B-NS3 interactions inhibits Zika virus polyprotein processing and protects mice from lethal challenge of Zika virus. Our current goal is to screen, identify, and characterize novel small molecule inhibitors against the viral protease, using both in vitro assays and in vivo animal models.

The flavivirus envelope (E) protein is a highly conserved and key target for developing therapeutic antibodies and vaccines. Although several neutralizing antibodies against the flaviviral E proteins have been identified demonstrated efficacy in preventing flaviviral infection in animal models, these antibodies are not ideal candidates as therapeutics due to Fc-receptor mediated antibody-dependent enhancement (ADE) effect. A single domain, termed VHH (variable domain of camelid heavy-chain-only antibody (HcAb)) or nanobody (Nb) has emerged as an effective drug candidate for various applications, including infectious disease management. VHH is the smallest intact antigen-binding fragment (15 kDa) harboring the full antigen-binding capacity of the original naturally occurring antibody and lacks the Fc domain to avoid ADE. The unique features of nanobodies make them an attractive potential mode of prevention and treatment for flavivirus and its associated complications. Our goal is to explore the potential of nanobodies as novel therapeutics for management of flavivirus infections.
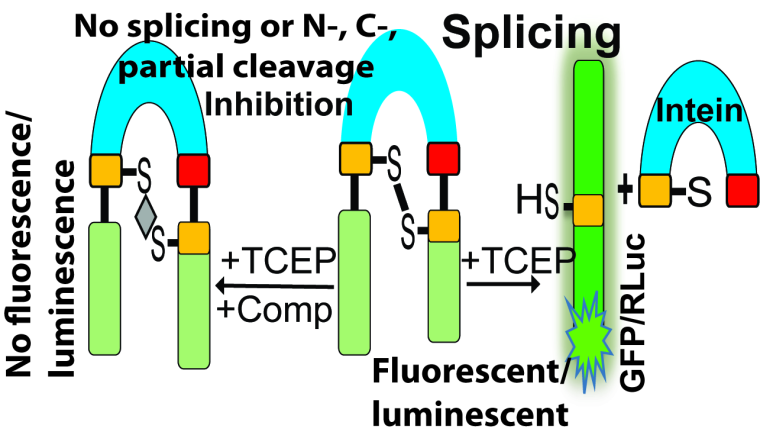
Many microbial pathogens contain protein self-splicing elements called inteins, which are internal protein elements that self-excise from their host proteins and catalyze ligation of the flanking sequences (exteins) with a natural peptide bond. Overall, more than 1,700 inteins have been identified. Certain intein-containing microorganisms are deadly human pathogens. For example, Mycobacterium tuberculosis (Mtb) has intein sequences in three critical genes involved in replication (DnaB), iron-sulfur cluster assembly (SufB), and recombination (RecA). Pathogenic fungi Crypotococcus neoformans (Cne) and C. gattii (Cga) have inteins in the genes encoding Prp8, an essential RNA splicing factor. The emergence of severely drug-resistant strains of Mtb and pathogenic fungi plus the deadly synergistic association with HIV/AIDS represent significant global challenges. Since inteins consistently disrupt the highly conserved sequences of host proteins, they often cause a disruption of functions that are essential for the pathogen’s survival. Because the inteins must self-excise from the precursor protein and splicing inhibition leads to the death of intein-containing microbes, inteins are attractive targets for drug development. Additionally, inteins do not occur in multi-cellular organisms and bacteria normal associated with the gut flora, making intein-inhibiting drugs highly selective only for intein-containing pathogens such as Mtb, Cne, and Cga. Our goal of this study is to determine the structures and functions of inteins from these pathogenic microbes, to develop high throughput screening assays to identify and characterize small molecule inhibitors targeting intein splicing activities.
